This project was completed towards the requirement of Stanford University’s BIOMEDIN 210: Modeling Biomedical Systems class. This project was completed jointly by Lichy Han, Winn Haynes, Maulik Kamdar and Alejandro Schuler. The PDF version can be downloaded here BMI210Report. The poster can be browsed here and the code is available online here.
Abstract
When performing bioinformatics analyses, finding similar projects can save a significant amount of time and effort. With more than 20 million repositories on GitHub, it can be daunting to find projects relevant to your particular research question. To address this challenge, we built a structured ontology of maintained bioinformatics packages which can be queried to find GitHub repositories which answer similar research questions. By developing this ontological structure, we are able to recommend projects to users which will provide sample code for developing their own software and recommend packages that the user may not have previously considered. We have distributed our search engine through a web interface which is available at http://tinyurl.com/biosearch-ui. From user evaluations, we found that users prefer our search engine to the default GitHub search in a 3:1 ratio. In creating this improved search platform, we hope to aid in the search and use of analytic tools to further biomedical research by improving efficiency, promoting collaboration, and encouraging innovation.
Background and Motivation
Recently, there has been an incredible movement towards public deposition of code, advocated by organizations such as the Open Source Initiative. Many source code repositories have grown to meet this demand, prominently including GitHub, SourceForge, and Bitbucket. GitHub, in particular, has over 20 million repositories and continues to grow at an exponential rate [Figure 1] [Doll 2013]. To further the complexity, each repository includes many source files. While the contents of GitHub span domains, these repositories contain thousands of different projects which may be of substantial interest to researchers in the bioinformatics domain.
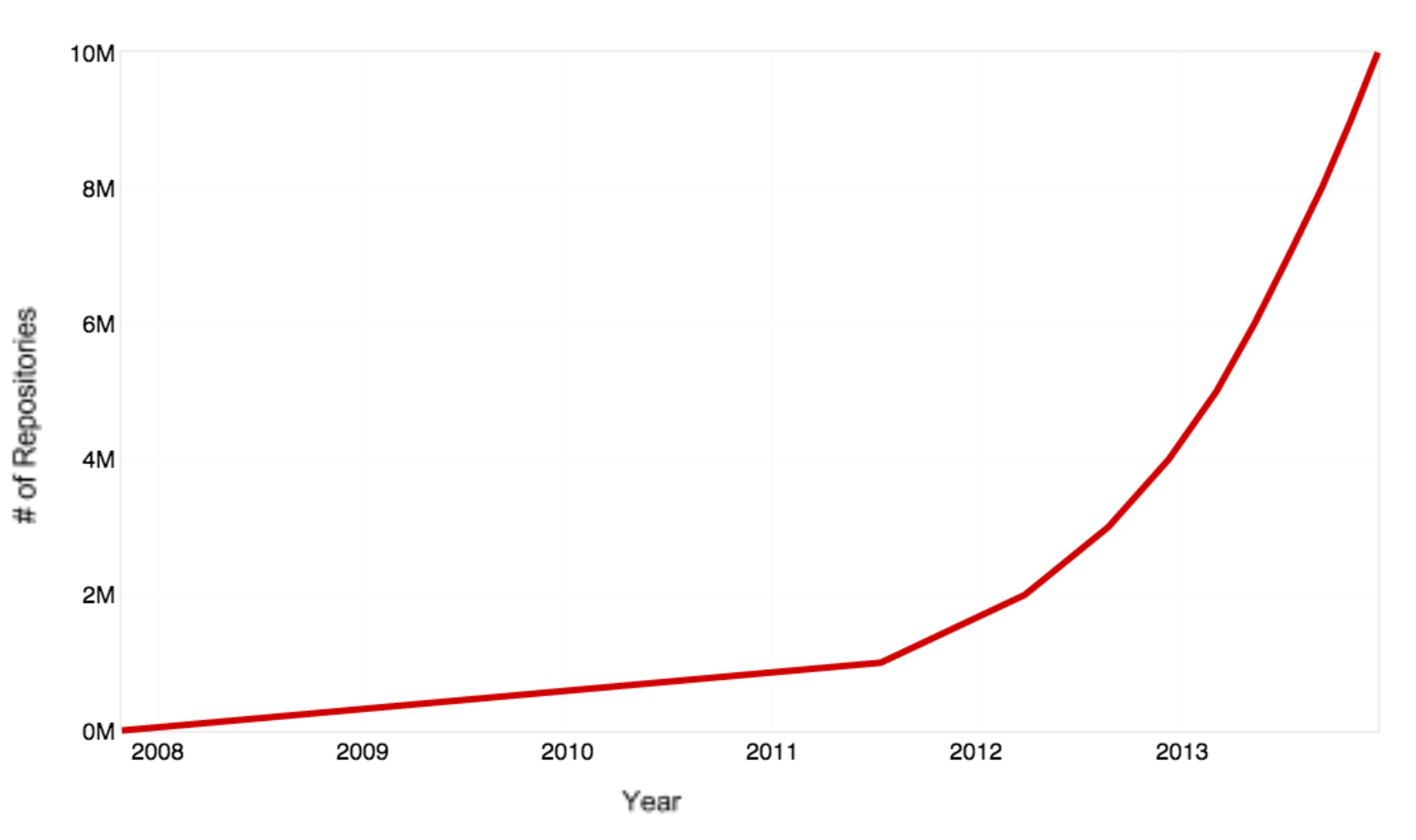 While this exponential growth rate presents a great opportunity to leverage the work of other scientists, a major challenge is effectively sifting through these vast amounts of source code. For instance, a simple query of “microarray” returns over 27,000 matching pieces of code. Fortunately, the bioinformatics domain relies heavily on shared software packages like Bioconductor and Biopython whose structure can be utilized to develop more effective queries. Bioconductor contains more than 2,000 packages which are developed for and maintained by active researchers in bioinformatics [Bioconductor]. The packages in Bioconductor are annotated with free text descriptions and classified into categories known as biocViews. biocViews are stored in a hierarchical structure, where node levels are based on the similarity of functional categories.
While this exponential growth rate presents a great opportunity to leverage the work of other scientists, a major challenge is effectively sifting through these vast amounts of source code. For instance, a simple query of “microarray” returns over 27,000 matching pieces of code. Fortunately, the bioinformatics domain relies heavily on shared software packages like Bioconductor and Biopython whose structure can be utilized to develop more effective queries. Bioconductor contains more than 2,000 packages which are developed for and maintained by active researchers in bioinformatics [Bioconductor]. The packages in Bioconductor are annotated with free text descriptions and classified into categories known as biocViews. biocViews are stored in a hierarchical structure, where node levels are based on the similarity of functional categories.
By using the semantic similarity of both the free text information and the hierarchical data, we prioritize GitHub packages which are of the most relevance to bioinformatics users with specific research questions. The development of these effective searching approaches for these open source code repositories enables utilization of prior work to improve efficiency, promote collaboration, and encourage innovation.
Methods
Ontology
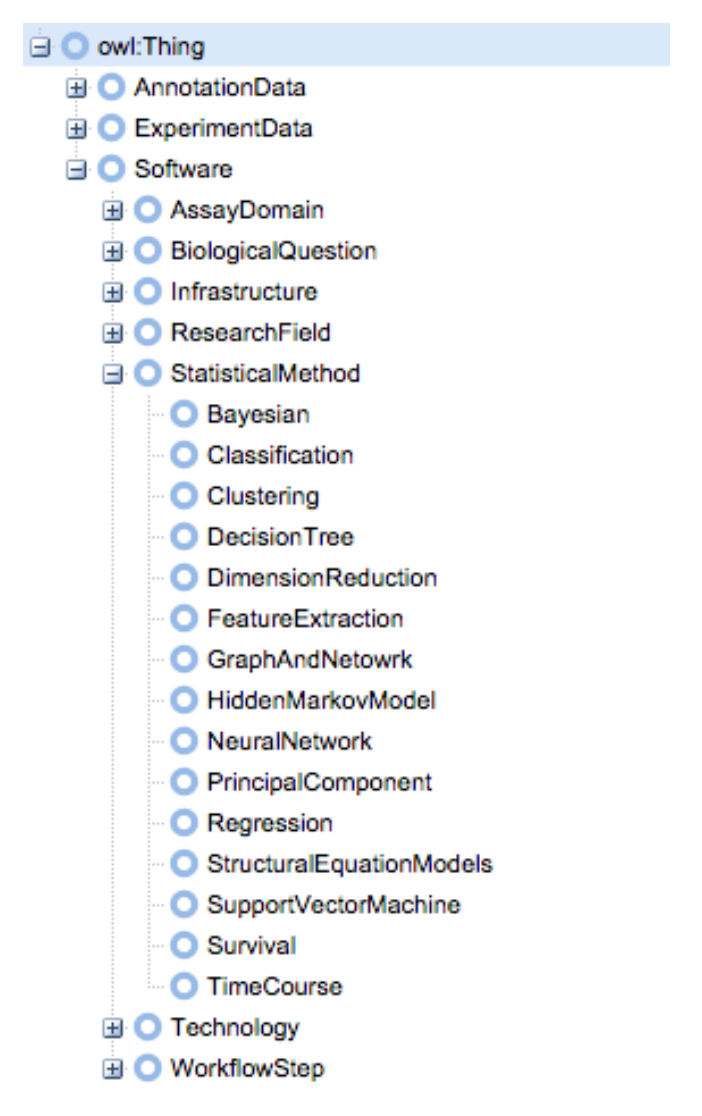 Using the existing hierarchy of packages in Bioconductor, we created an OWL-based ontology using WebProtégé [Figure 2] [Bioconductor]. WebProtégé allowed us to collaboratively develop the ontology through an intuitive web interface [WebProtege]. Our ontology is available at http://tinyurl.com/biosearch-onto.
Using the existing hierarchy of packages in Bioconductor, we created an OWL-based ontology using WebProtégé [Figure 2] [Bioconductor]. WebProtégé allowed us to collaboratively develop the ontology through an intuitive web interface [WebProtege]. Our ontology is available at http://tinyurl.com/biosearch-onto.
At the top of the ontology, we have the three general classes of packages: annotation data, experiment data, and software. Software packages are the main focus of our project, and include tools for analyzing a wide variety of experimental data. As an example, if we were interested in performing a time course analysis, we would look at the “TimeCourse” node which is a subclass of “StatisticalMethod” and “Software”.
For each package, we extracted the associated annotation data from Bioconductor. Specifically, using Python, we extracted the “biocViews,” which are keywords that Bioconductor assigns each package and served as the basis for the construction of our ontology. In total, we extracted data for 2,058 packages relevant to our hierarchical model. For instance, “affycoretools” is annotated with the classes “GeneExpression”, “Microarray”, “OneChannel”, “ReportWriting”, and “Software”.
In addition to the structured component of the Bioconductor package annotations, we pull the free text portion of the descriptions to identify relevant packages based on unstructured term frequency. We utilize this information to provide context for package utility beyond the limited structured vocabulary enabled by the biocViews ontology.
Problem-Solving Methods
We used the GitHub API to download the metadata of the repositories whose source code is written in R and import any package from Bioconductor [GitHub]. As the GitHub API does not allow a ‘blind’ code search over all its repositories, we decided to build a list of potential GitHub repositories which could be related to bioinformatics and clinical informatics research. We generated a bag of words from the descriptions of the Bioconductor packages and retrieved the metadata of all those repositories which mentioned any of the words in the repository name, description, or Readme. We extracted the following information from the API response: id, name, full_name, score, update_date, create_date, html_url, watchers_count, stargazers_count, forks_count, language, size, description and whether it is a private repository. This metadata information was stored in a PostgreSQL table. To increase the speed for searching each package import across these repositories (GitHub API limits to 20 requests per minute), we downloaded the source code in the ‘Master’ branch of these repositories to a local server. The package-repository associations were stored in a PostgreSQL table, with the score of the association, and the files where the package was imported.
After a user enters her query, our system performs a three-step information retrieval process in order to present the user with a rank-ordered list of GitHub projects relevant to her needs [Figure 3].
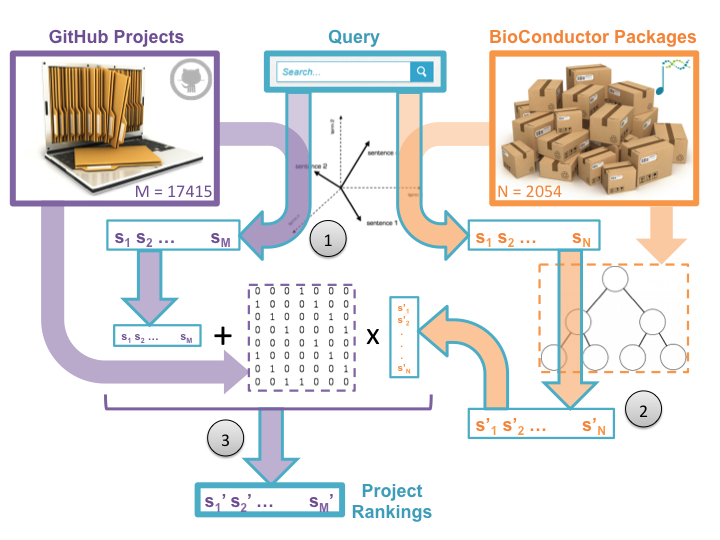
In the first step, we score each Bioconductor package and GitHub project by relevance to the users’ query. The system assigns these scores by calculating the cosine distance of the query’s term frequency vector to precomputed term-frequency inverse-document-frequency (TF-IDF) indices of the package annotations and project descriptions [Salton and McGill, 1983]. The result is two sets of scores: one set for Bioconductor packages and one for GitHub projects. These scores reflect the textual similarities between the query and project and package documentation.
In order to leverage our knowledge of Bioconductor packages, we subsequently perform a type of diffusion of the package scores based on semantic similarity (step two). In our implementation, this is accomplished by multiplying the package score vector by a row-normalized inverse semantic distance matrix, where the semantic distance between two packages is defined by the length of the shortest path between them in our hierarchical ontology [Schickel-Zuber and Fallings 2007, Resnik 1995, Leacock and Chodorow 1998]. Sharing scores over semantic distance means that packages which are ontologically “close” to high scoring packages may have their scores boosted as well, even if their annotations do not include terms in the query. Similarly, packages that score highly but are not ontologically similar to other high scoring packages may have their scores attenuated. This intelligent behavior naturally mitigates the effect of poor or inaccurate package annotations.
In the third step, we combine the smoothed package relevance scores with the GitHub project relevance scores to create a rank-ordered list of projects to suggest to the user. We calculate each project’s final relevance score as it’s project-to-query similarity score added to the smoothed package relevance scores for every package used in the project. This leverages our BioConductor domain knowledge to intelligently boost the scores of projects that use known-to-be-relevant tools, even if the project is poorly annotated and would fly under the radar of standard textual search.
After sorting, the final result of this process is a detailed list of GitHub projects that is rank-ordered by a smart measure of the relevance of the project to the user’s query.
Evaluation
Search engine evaluation is inherently a difficult process [Vaughan 2004, Chu, et al 1996]. The primary method for making quantitative assessments is having experts determine the relevance of each potential result against testing queries and run those queries against the search engine. By comparing top-ranked results to the expert indicated relevant entries, measures of accuracy can be calculated. In addition to being logistically difficult, quantitative evaluation fails to capture users’ intuitions in creating queries or their natural iterative interaction with the engine. In the future, we hope to acquire the resources to enable quantitative testing, but in our current scope we focus on understanding qualitative performance.
To evaluate the qualitative performance of our system, we amassed a group of 14 test users and asked them to fill out a qualitative assessment. We selected a diverse group of biosciences researchers to better understand how our system fared depending on the needs and backgrounds of the users.
Our survey displayed results side by side from GitHub’s search engine and our search engine. Survey responders were blinded to which underlying search engine was used. The top five results were displayed from each search engine, and the order that the group of results were shown in was randomized for each set of questions. Users were then asked to rate the relevance of top results from both engines and select which search engine they preferred overall, where GitHub’s search engine was labeled as Search Engine A and ours as Search Engine B. We provided three scenarios in the survey, and compared the search engines using multiple choice, short answer, and Likert scale questions.
The questions in our survey, which can be accessed at http://tinyurl.com/biosearch-survey, included questions of the form:
- On a scale of 1 to 10, overall, how would you rank the results from Search Engine A?
- Which search engine’s results do you prefer, A or B?
- On a scale of 1 to 10, how relevant are the following results from Search Engine A to the query?
- What are some ways you might change the query to improve these results?
- If you were to use our search tool, what would you be likely to use it for?
- Write a query to search for your software need.
The majority of the questions were marked as mandatory to ensure that users carefully considered each question and response. Questions of the first type elucidate in a more unbiased fashion what the usefulness of our system is in retrieving results that are relevant to arbitrary queries. The free response questions of the second type help us understand the user’s thought process and intuition when interacting with the system.
Results
Application Prototype
During the first iteration, we implemented our application as a Python-based script where the user provided a search term and the number of results to be displayed as arguments to the script in the command line console. The script executed the three-step information retrieval process, as described previously, and returned a ranked-list of GitHub repositories which were the most relevant to the user query. While effective, it required the users to store the scripts and the TF-IDF index locally to use our search system.
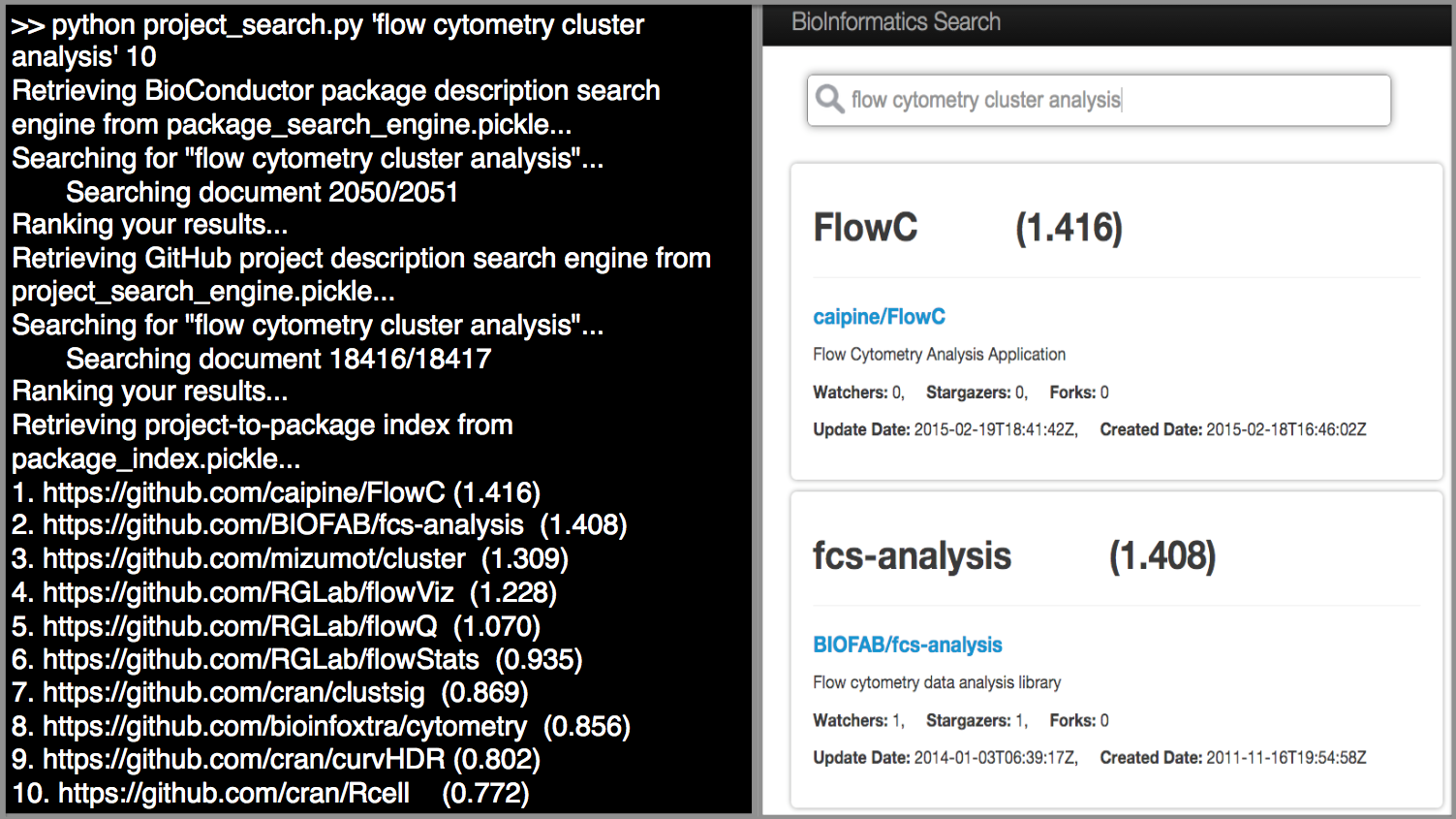
Hence we developed a Web-based Search Graphical User Interface (GUI) to mitigate this challenge, and provide a one-stop interactive search experience for the user [Figure 4]. We deployed the python scripts, the TF-IDF index and the PostgreSQL database containing the metadata of the GitHub repositories on an Amazon EC2 instance, along with the Search GUI on an Apache 2 Server. The user types his query in the search box, which invokes the commands in the Python script to retrieve a list of top 30 most relevant projects. Each GitHub URL identifier is passed as a parameter for an SQL select query, to retrieve the metadata of the repository – the modification and creation dates, owner and number of forks, watchers and stargazers. This set of results are presented to the user, where he can directly open the GitHub repository. The Search GUI is made available at http://tinyurl.com/biosearch-ui.
Evaluation Results
As described in the Methods above, our evaluation was based primarily on a survey comparing results from our search engine to the native Github search engine. In total, we compiled survey responses from 14 volunteers [results available at http://tinyurl.com/biosearch-results].
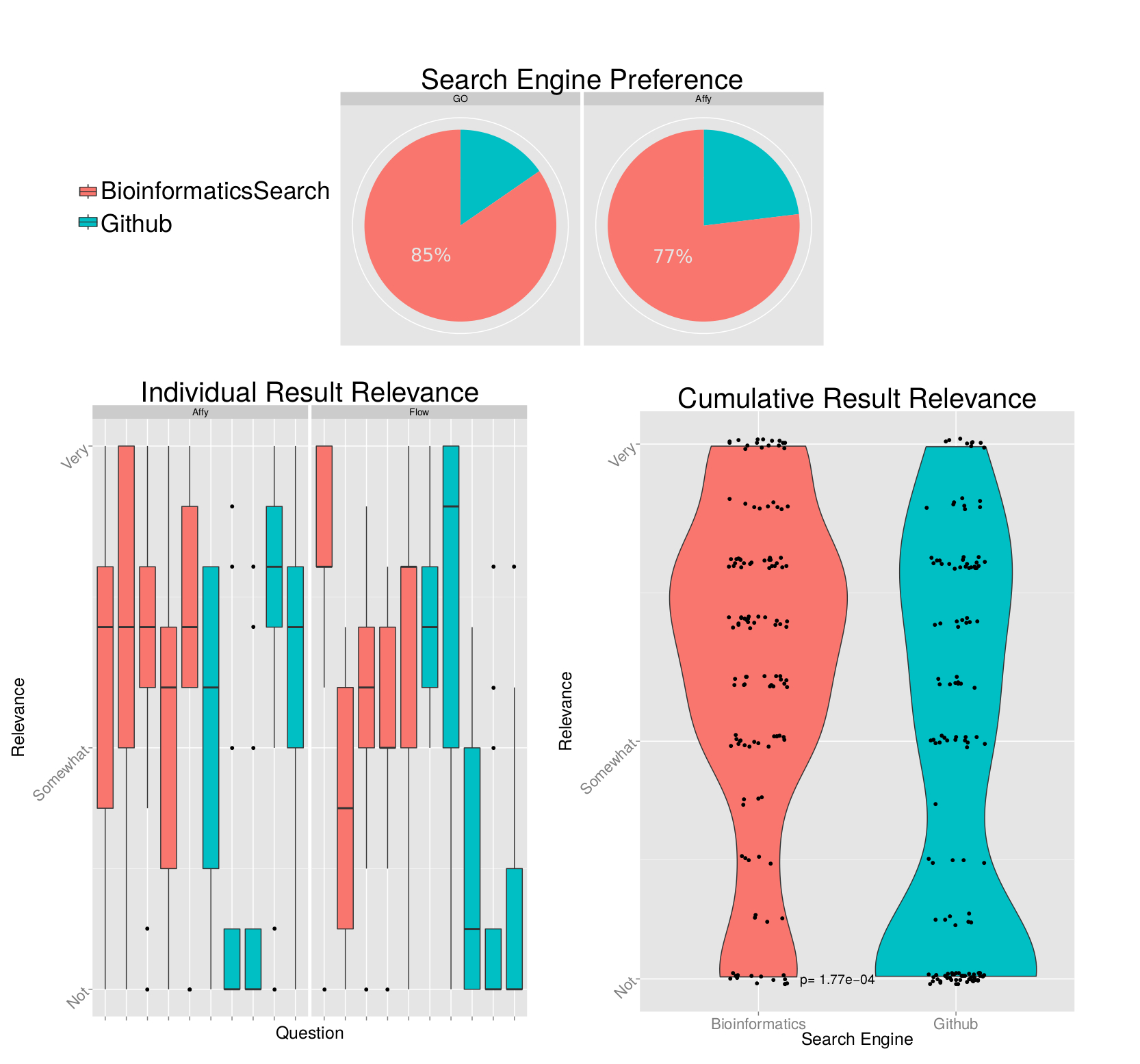
We asked respondents to rate their overall preference towards the different search engines based on several different query results. We found that users preferred our search engine to Github’s search engine on all test queries, in roughly a 3:1 ratio [Figure 5, top]. Please note that we elected to not include results from our sample query related to flow cytometry because our wording of the question led to confusion and inconsistencies in respondent answers (Search Engine B was listed before Search Engine A).
For some of the sample queries, we asked users to rate the individual relevance of the top 5 results from each of the search engines [Figure 5, bottom left]. We asked users to rank their relevance on a scale of 1-10, where 1 is not relevant and 10 is very relevant. We observed that results returned by the default Github search engine are highly variable: some results will be quite relevant, while others are consistently reported as having no relevance. In contrast, our search results are more consistently ranked as relevant.
Finally, we combined the rankings for all of the individual relevance rankings by search engine [Figure 5, bottom right]. From a qualitative perspective, we observe that the Github search engine has a much higher density of respondents ranking results as “not relevant.” To quantitate the differences in these distributions, we performed a Wilcoxon rank sum test between the distributions and calculated a p-value of 1.77e-04, which would pass all but the most stringent of statistical significance filters. We selected the Wilcoxon test because we did not want to make any assumptions of normality in these distributions.
Discussion
The primary contribution of our project is the ability to find code to address a user’s need even if that code has not been properly documented, or to provide additional confidence in the code’s utility if it is well annotated. However, the methods described here could be extended for developing applications to semantically search across documents and unstructured datasets in other domains. Our evaluation results indicate that the search recommendations generated by our system are more relevant to the user queries than a blind text search across GitHub, demonstrating the power of ontologies in Search.
Future Work
In this project, we show how searching across GitHub repositories importing Bioconductor packages could be improved to provide the user with the most relevant results. This could be extended vastly to include other languages and bioinformatics-specific modules (BioPython, BioPerl etc.) in these languages. Our ontology could be made more contextually rich by incorporating axioms on package functions and classes, languages, and classifications provided by other modules. Our hierarchical ontology could also be improved by creating additional levels of biological intuition. In particular, the maximum depth of our hierarchy is presently five levels, a number which could be substantially increased with further curation.
There are also several ways that the information retrieval process could be improved within our framework. A simple improvement would be to stem or lemmatize the words in the documentation and query so that inexact queries would still match relevant documents, e.g. “semantic” would match “semantics”, “analyzing” would match “analysis”, etc. In addition, the semantic smoothing of package scores currently rests on an overly-simplistic measure of semantic similarity. Researchers have suggested alternate methods that could improve the relevance of our results [Schickel-Zuber and Fallings 2007, Resnik 1995, Leacock and Chodorow 1998]. Using a richer representation of semantic distance would not retard runtime performance because the distance matrix is precomputed based on the structure of the static ontology.
The computation of the final project score could also improved by including measures of project popularity and better normalizing the included package scores. The former could be implemented by producing a score based on the number of stars or watchers on the project, while the latter could better account for the distribution of package scores for a given query. Other measures also include TF-IDF scores over the GitHub issues and user comments on each commits and files, which we already retrieve. These features would not be guaranteed to improve overall performance, so their inclusion would have to be rigorously evaluated.
The Search GUI could be improved upon to provide a richer user experience. More specifically, as primary steps, we envision that features like Type-ahead Autocomplete, where the user is prompted with options as he types in his query, and Faceted Search, where he can filter the results based on the language of the projects, forks, issues, modification dates etc. and user personalization could be provided. An ability to access the GitHub page from within the GUI (frames or screen grabs) could also be implemented, so the user could summarily browse the code before actually visiting the page. User experience and usability could be evaluated using a silent observational methodology like Tracking Real Time User Experience (TRUE) [Kim et al. 2008].
References
Bioconductor. (n.d.). Retrieved February 27, 2015, from http://www.bioconductor.org/packages/release/BiocViews.html#___Software
Chu, H., & Rosenthal, M. (1996). Search Engines for the World Wide Web: A Comparative Study and Evaluation Methodology. Proceedings of the ASIST Annual Meeting, 33, 127–35. Retrieved from http://www.editlib.org/p/83947/
Doll, B. (2013). 10 Million Repositories. Retrieved from https://github.com/blog/1724-10-million-repositories
GitHub. (n.d.). GitHub API v3. Retrieved February 27, 2015, from https://developer.github.com/v3/
Kim, J. H., Gunn, D. V., et al. (2008, April). Tracking real-time user experience (TRUE): a comprehensive instrumentation solution for complex systems. In Proceedings of the SIGCHI conference on Human Factors in Computing Systems (pp. 443-452). ACM.
Leacock, C., & Chodorow, M. (1998). Combining Local Context and WordNet Similarity for Word Sense Identification. An Electronic Lexical Database, 265 – 283.
OSI. (n.d.). Open Source Initiative. Retrieved February 27, 2015, from http://opensource.org/about
Resnik, P. (1995). Using Information Content to Evaluate Semantic Similarity in a Taxonomy, 6. Computation and Language. Retrieved from http://arxiv.org/abs/cmp-lg/9511007
Salton, G., & McGill, M. J. (1983). Introduction to modern information retrieval. Retrieved from http://agris.fao.org/agris-search/search.do?recordID=US201300296398
Schickel-Zuber, V., & Faltings, B. (2007). OSS: a semantic similarity function based on hierarchical ontologies, 551–556. Retrieved from http://dl.acm.org/citation.cfm?id=1625275.1625363
Vaughan, L. (2004). New measurements for search engine evaluation proposed and tested. Information Processing & Management, 40(4), 677–691. doi:10.1016/S0306-4573(03)00043-8
WebProtege. (n.d.). Retrieved February 27, 2015, from http://webprotege.stanford.edu/#List:coll=Home;