As part of our requirement towards successfully ‘graduating’ from the 10th Summer School of Ontology Engineering and Semantic Web (SSSW 2013), we were required to complete one Mini-project. By the definition of the mini-project, different teams were given specific research problems, which were to be tackled within a short duration of 4 days, under the guidance of a mentor who initially proposed the problem. The research problems could be both – application-based or something that required a more theoretical perspective. Tackling it was difficult, not only because of the short span of 4 days, but even during those 4 days, there were various invited talks, hands-on sessions and different social events on each day (Trip to Segovia, Swimming Pool etc.), which should never be missed. Moreover the team had to come up with a humorous video to present during the Gala Dinner on the final night – hence, the project itself validated some long nights.
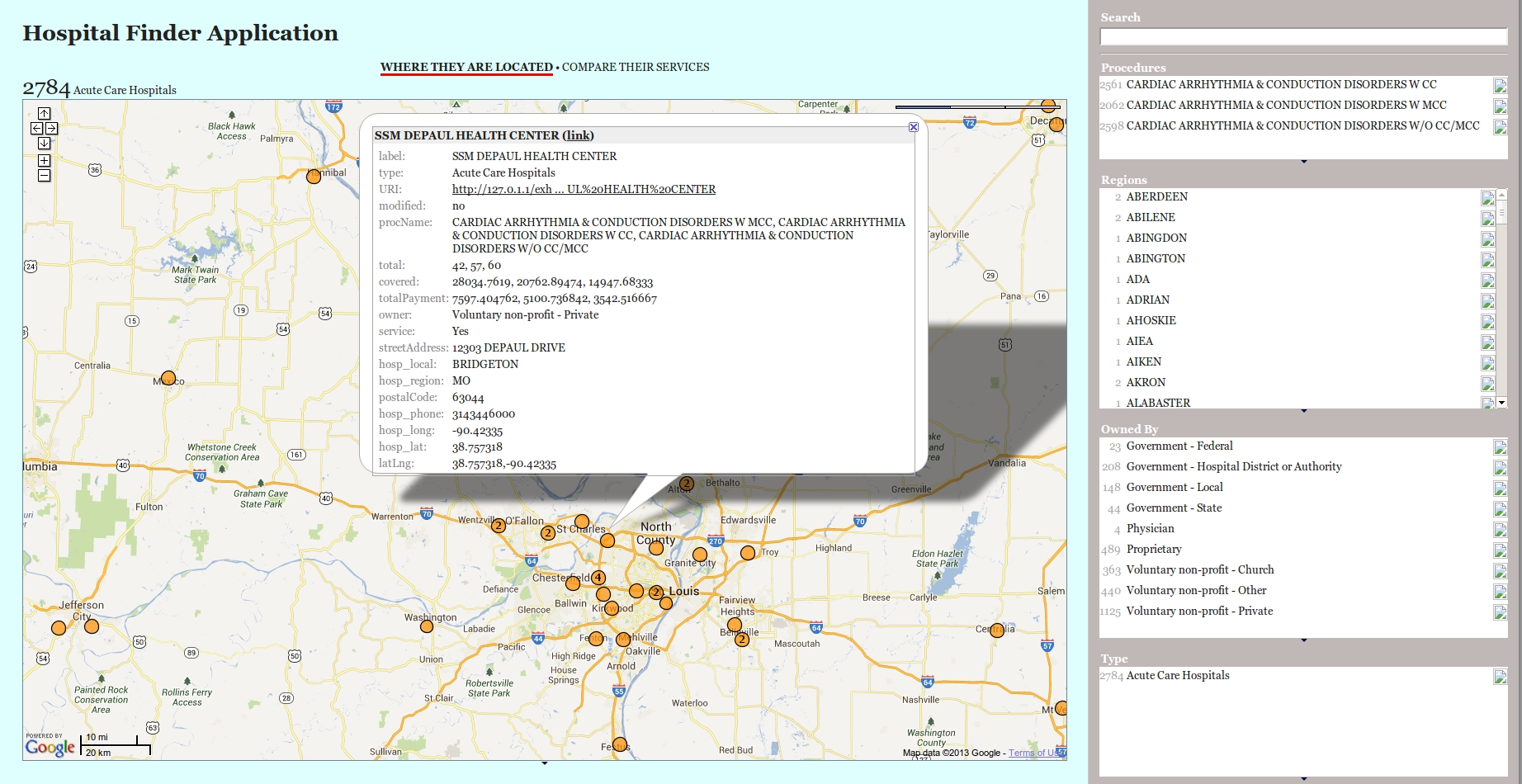
We were presented with the development of a Hospital Finder Application by integrating various bio-medical data sources, containing information relevant to the Hospital Demographics, their Inpatient Procedure Rates (surgical procedures which were insured), Outpatient Calls (Ambulatory Processes), Hospital Quality Data from Medicare, Clinical Trials data, etc. The idea was to develop an application (or present an approach) which would allow any user to search the disease, he is suffering with, and the application pushes out a list of procedures responsible for its effective treatment (inpatient or outpatient) and which hospitals provided the desired services. He could then visualize the hospital locations and also compare the different rates and the hospital quality. We decided to use a faceted browser (namely Exhibit), where he could then drill down according to his requirements. However to reach to that stage, we had to tackle the mammoth challenge in front of us – the integration of these data sources. For instance, the inpatient data source listed the procedures against the hospitals carrying them, but it was impossible to determine, which disease or ailment required the procedure. Moreover the procedure codes in this data differed in format from those outlined in the ontologies listed under Bioportal. Hospital demographics presented the accurate address and US postal codes, but it was not possible to find the Latitude and Longitude information, something that was required to accurately pin the location on a Google Map Visualization. Another problem was that these data sets had to be standardized to a common format for exchange, and currently, they were either in CSV or TSV files, or worse as Oracle Dumps. It was lucky that the the different data sources we planned to integrate (apart from Clinical Trials) had a single unique column, pertaining to the ‘Provider ID’, a Unique Hospital Code which remained consistent across them.
We started off with the pre-processing and refining these datasets using Google Refine RDF Extension, which allows to semantically annotate any data-set using an imported schema, as well as perform naive entity reconciliation with DBPedia or Freebase Entities (or you can specifiy your own SPARQL Endpoint) using the rdfs:label property values. Google Refine allows import of the data files in varied formats, and we re-used the properties available from Schema.org, namely schema:name, schema:address, schema:postalCode, schema:addressRegion, schema:streetAddress and schema:addressLocality alongwith some custom-engineered properties to facilitate the linking. The model graph is shown below. The idea behind this modelling of the data sources was that every hospital (identified by Provider ID) conducts a set of procedures, which could be used to treat certain diseases (naive string matching against procedure name). Moreover, the hospitals would also provide certain outpatient emergency procedures (where the patient is not admitted). The hospital is identified by a street address, which was interlinked with the appropriate geographic coordinates (Latitude, longitude) using GeoNames by translation of Postal Codes. An approach was shown towards entity reconciliation against DBPedia Sparql Endpoint for extra information about the hospitals and places. These RDFized datasets were stored in a Sesame Repository.
Due to my familiarity with the use of Exhibit and the concepts of faceted and lens-based browsing, we quickly decided to conjure up a working prototype with a use-case on Cardiac Arrhythmia – a person suffering from this disease tries to search nearby hospitals which perform the surgical procedures relevant to it, with the condition that the hospital is somewhere nearby and is under the desired price range. Due to the network issues in Cercedilla, our hope was that we could put forth a convincing demo, showing how the user navigates through our interface and makes an informed decision.
We finally decided to present the Mini-project, by starting with a video, showcasing our problem in lucid terms, and including a screencast of the initial prototype that we had designed. This video can be seen below :-
Our mini-project won the best project award out of 11 teams (there were no official prizes given, but just three best project awards), and all the team members received a USB Microphone (my third USB Microphone after the Best Poster Award, and 2nd Best Video!). It was really fun, and I got a lot of insight into data integration, biomedical ontologies and modeling.
Our team from right – Antonio, Bjarte, Dr. Natasha Noy from Stanford, who supervised the entire project, Nadin, me and Yves and Dr. Sean Bechhofer, who organized the mini-projects at the prize distribution ceremony.