Proposal : https://maulik-kamdar.com/2011/05/the-reactome-project-proposal/
Demo : http://reactomedev.oicr.on.ca:8080/ReactomeCanvasBrowser/ReactomeBrowser.html
Acknowledgement: SourceForge
During the end of April 2011, I was selected for my second Google Summer of Code Program. My project this year combined my two fields of interests, as I had always wished for – Biotechnology and Data Visualization. I was supposed to work for the next 4 months for the Reactome Project under the organization Genome Informatics, a group organizing the joint efforts of WormBase, Reactome, and GBrowse.
The Reactome Project centrally consists of a manually curated database of core pathways and reactions in human biology that functions as a data mining resource and electronic textbook. The Reactome data model describes diverse processes in the human system, including the pathways of intermediary metabolism, regulatory pathways, signal transduction, and high-level processes, such as the cell cycle. Reactome software uses only freely available (and often open source) components and has been created with cross-platform compatibility and wide usability in mind. Data is stored in a MySQL database, the web site is implemented in Perl and data entry tool in Java programming language. The Reactome team is composed of individuals who are both biologists and programmers at the Ontario Institute for Cancer Research, New York University Langone Medical Center, Cold Spring Harbor Laboratory, and The European Bioinformatics Institute.
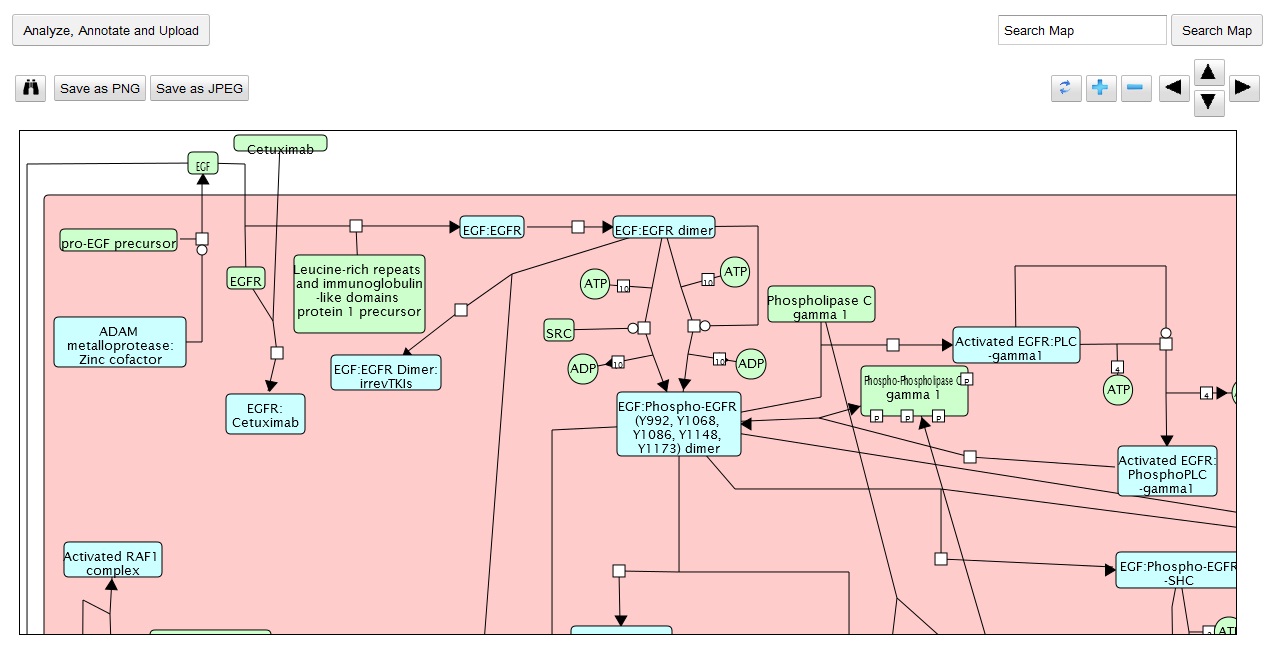
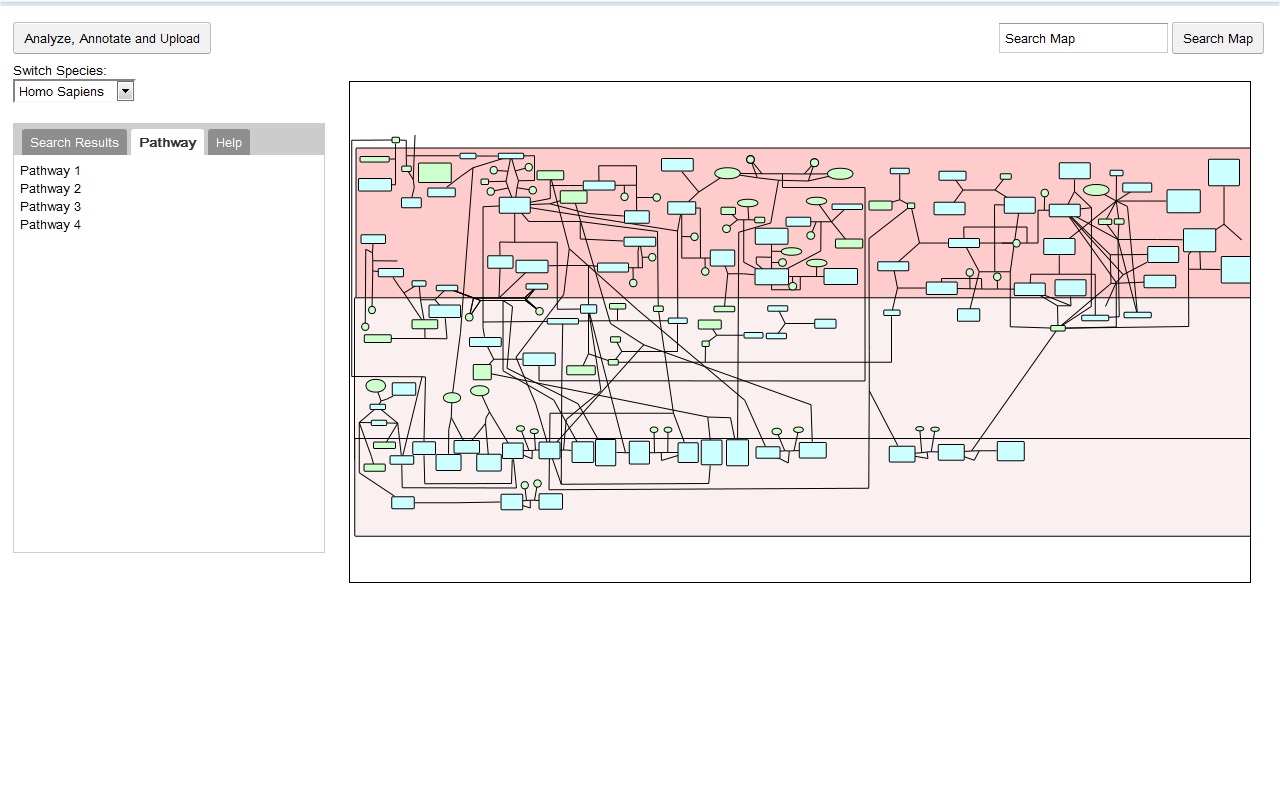
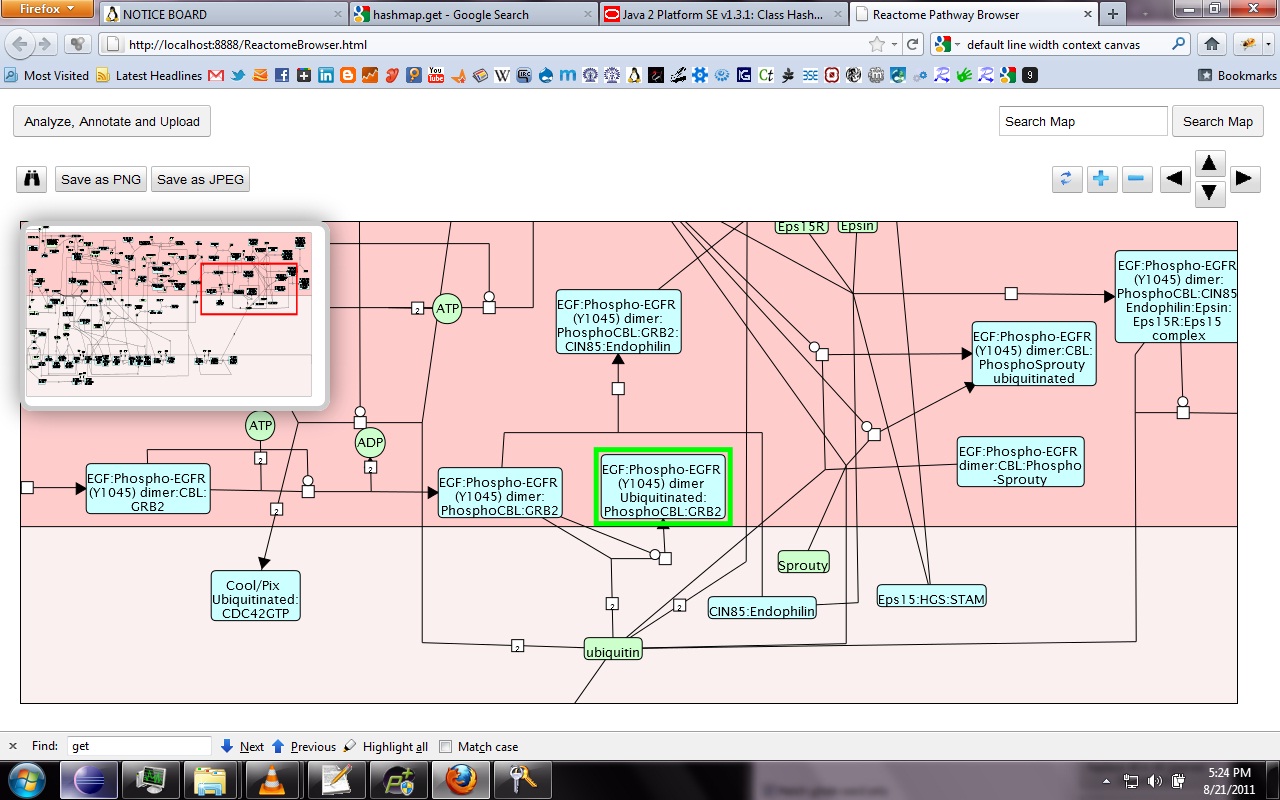
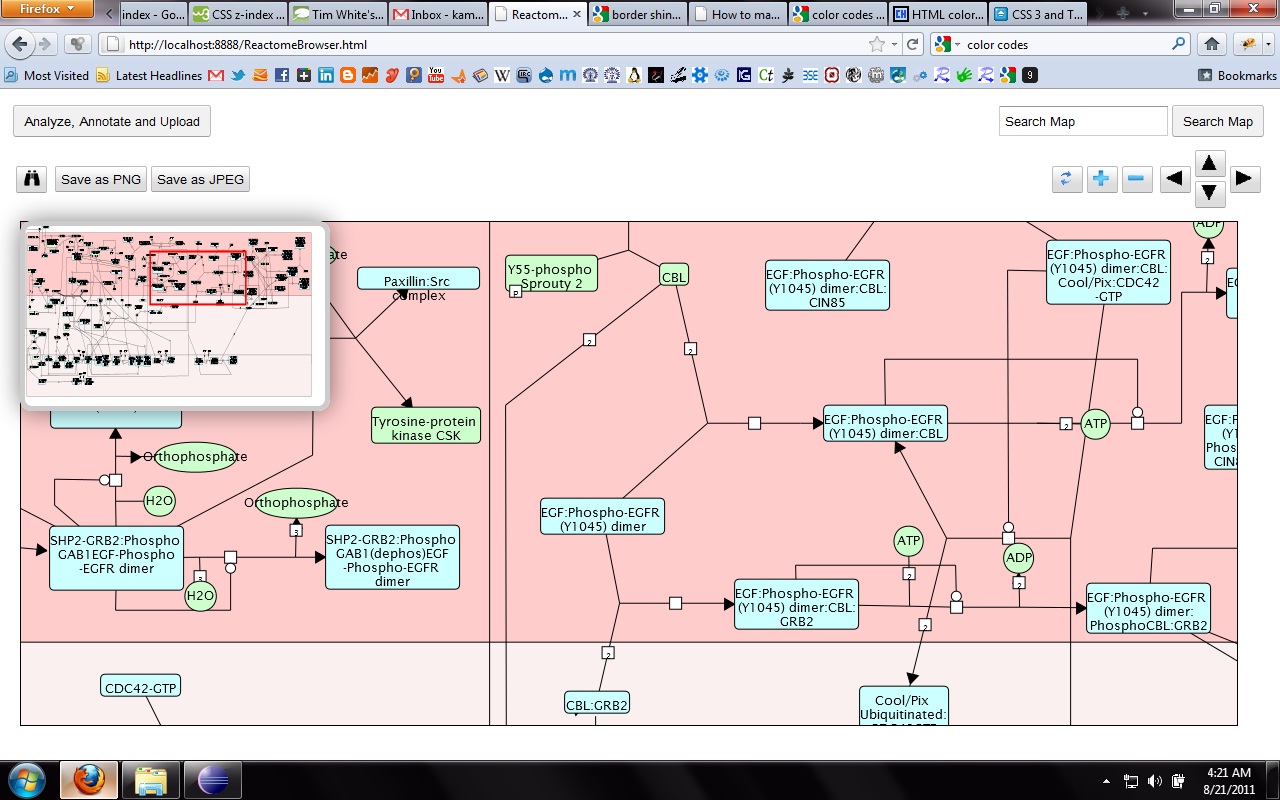
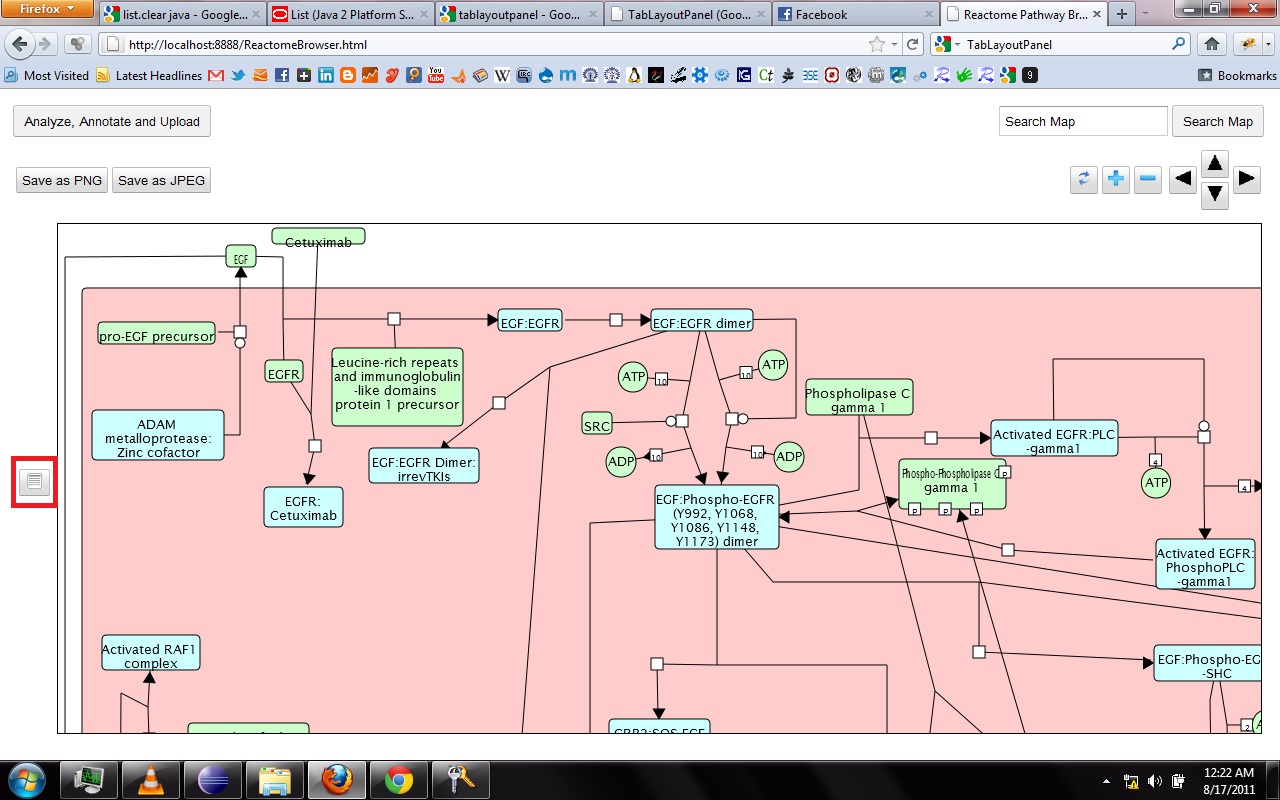
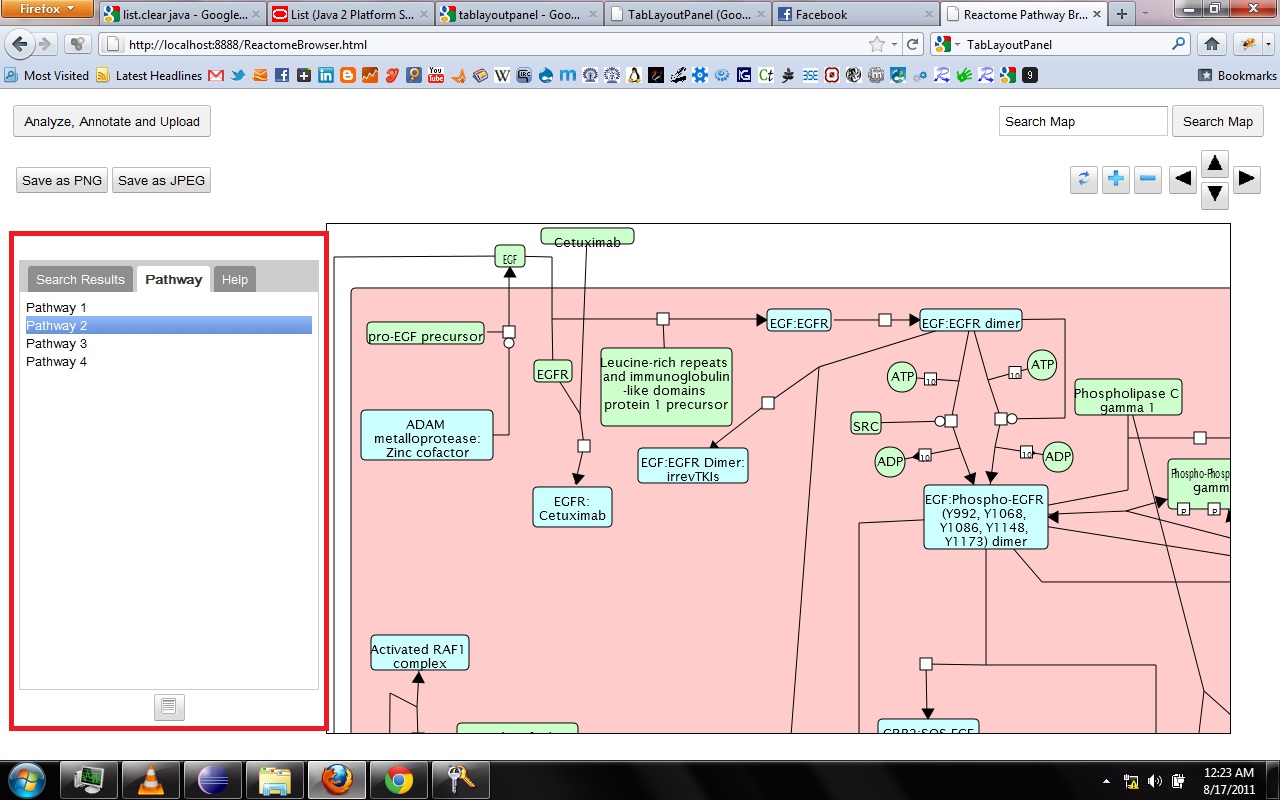
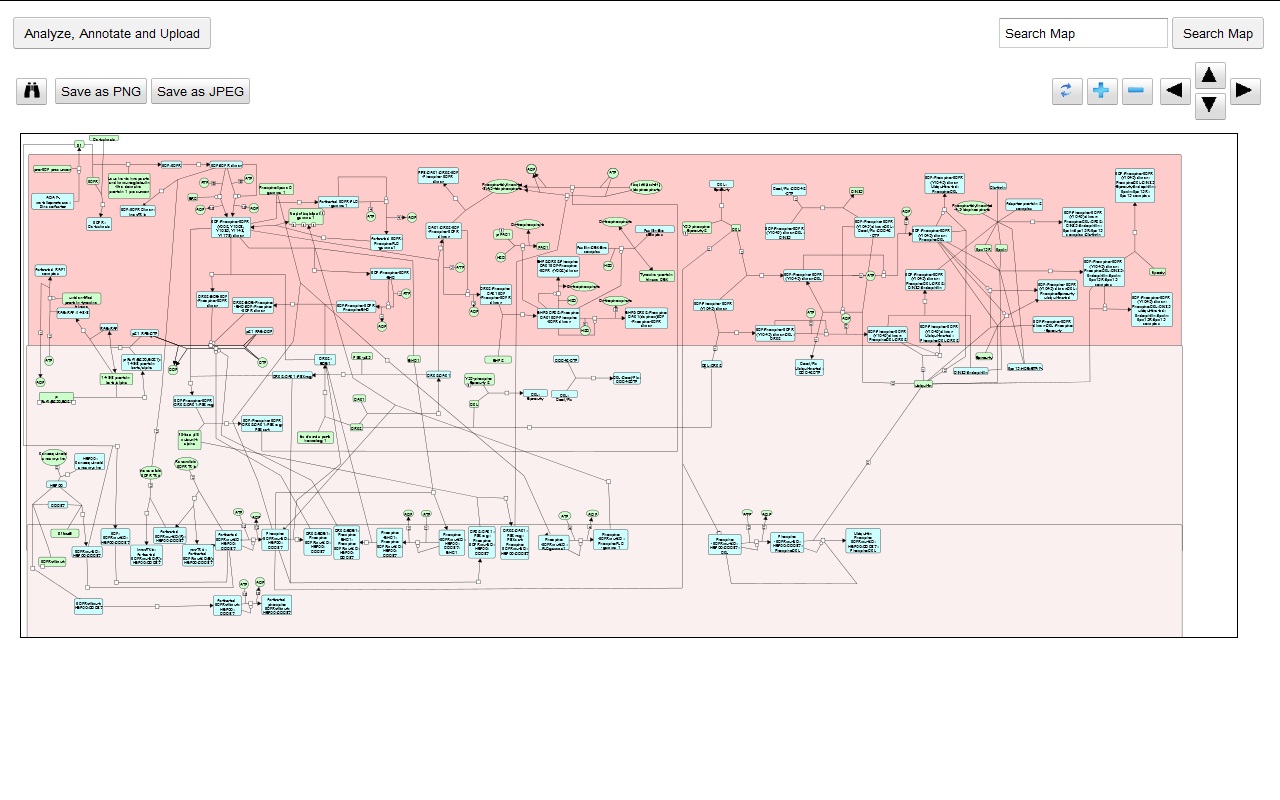
My main goals revolved around the development of a Reactome Pathway Browser, a Systems Biology Graphical Notation (SBGN)-based interface, based on the HTML 5 Canvas, that supported zooming, scrolling and event highlighting, for the visualization of curated Biological Networks & Metabolic Pathways. The Browser fetched data from the Reactome Database, in XML Format through RESTful Communication. It then parsed the huge XML files, holding datasets of the nodes and edges of manually curated Metabolic and Biological Pathways, and subsequently plot on the canvas using this parsed data. I added a few Canvas handlers to increase the interactivity and user experience. I have developed the system to later allow real-time multi-researcher collaboration to simultaneously edit any pathway, and track revisions.
The final product would replace the traditional Image based Browser used for the visualization of the Pathways. It would serve as an important Bioinformatics Tool and I am currently testing it for the addition of new features, as well as for official release purposes. The code is completely Open Source and is stored on Google Code Repository.
Here are few of the developmental screenshots of my project Reactome Canvas Browser. I would soon be sharing my notes and few code snippets, about how I have gone about developing the functionality. The project would hopefully soon see its dev release for testing purposes.