Proposal : https://maulik-kamdar.com/2012/04/reactome-pathway-summary-visualization/
Code : https://github.com/maulikkamdar/reactomepathwaysummary
Demo : http://reactomedev.oicr.on.ca:7080/reactomepathwaysummary/ReactomePathwaySummary.html (Non-functional due to removal of support of the underlying REST API – Check the images/Wiki)
Wiki : https://github.com/maulikkamdar/ReactomePathwaySummary/wiki/Reactome-Pathway-Summary-Visualization-Application
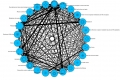
Since March 2012, I had been working on the development of a second Canvas Browser for the Reactome Project. The purpose of this browser would be to provide an overview of all the pathways in the Reactome Database. Top-level pathways are laid out in circular or force-directed layouts as circular nodes, and are connected to neighboring pathways (nodes) depending on the relationship shared between these two pathways. The user can navigate across the layout and gain an insight of the total number of participating molecules in any particular pathway which is in a direct proportion to the size of the node. The relationship between two pathways is decided by an algorithm which assigns different weights to parameters like the common number of participating molecules between the pathways, reactions shared between pathways etc. and generates a similarity measure which determines the thickness of the edge connecting the two nodes. All this information is fetched via FrontPageItems and PathwayParticipants REST API. A threshold value could be set via a slider, to visualize only those edges which have a similarity value above the given threshold value. A small video demo for the force directed layout can be found here. A hierarchical layout is also provided along with the circular and force-directed layouts. The user can visualize second degree pathways arranged in a radial sense. The hierarchical layout however does not provide any information about the total pathway participants in a pathway node.
Moreover users can now visualize their Expression Analysis as well as Species Comparison Analysis Results using the Pathway Summary Browser. The results could be in percentage format or in simple numeric format (differential expression). The degree of expression over the pathways is mapped to a color scale and the color of the nodes is adjusted accordingly. To facilitate users with the Red-Green Color Blindness the color scale currently has been kept as gradients of yellow (negative values) to blue (positive values).
Some screenshots of the browser are attached below.